
- Home
-
Services
 Leading molecular discovery services
Leading molecular discovery services
 Antibody Discover Service
Antibody Discover Service
 Technology platform
Technology platform
 Technology optimization service
Technology optimization service
- Products
- Resources
- Publication
- Contact Us
Yeast Surface Display Platform
Principle of Yeast Surface Display Technology
Yeast Surface Display (YSD) is a technique that presents foreign proteins on the surface of yeast cells. The process involves fusing the gene encoding the foreign protein with a yeast surface display vector, which is then introduced into yeast cells. This enables the expression of the foreign protein on the yeast surface, allowing the display of various biomolecules such as antibodies, proteins, peptides, enzymes, and others.
Yeast surface display mechanism
Our Yeast Surface Display Platform utilizes an α-agglutinin display system. In this system, the VHH gene sequence, along with an HA tag expression sequence, is inserted into the Aga2 subunit gene of the protein scaffold vector. The vector is designed with a nutritional marker to maintain selective growth in yeast. By adding galactose secreted into culture medium and anchored to the yeast cell wall and bonded to Aga1p subunit through two disulfide bonds (Figure 1).
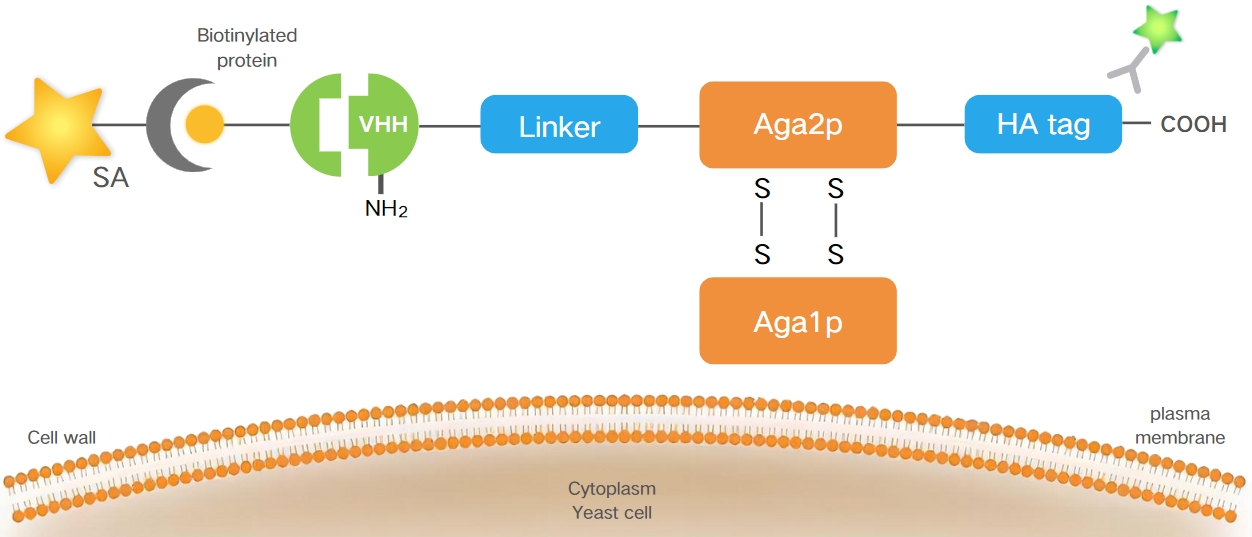
Figure 1. The VHH gene and HA tag are inserted into the Aga2 subunit,, and the VHH-Aga2p fusion proteins are anchored to the yeast cell wall via disulfide bonds to Aga1p.
Yeast Surface Display Library Construction
Our platform ensures exceptional library quality through rigorous validation processes:
1. Primer Optimization: We use proprietary primers designed to comprehensively cover all HcAb germline sequences.
2. Zero-Background Cloning: By employing background-free cloning technology, we guarantee 100% VHH insertion efficiency.
3. Library Capacity: We guarantee a library size of over 1×10^8 CFUs.
4. Clonal Uniqueness: After randomly selecting 48 clones for Sanger sequencing, we ensure 100% clonal uniqueness with no sequence duplication detected.
5. Sequencing Depth Analysis: We guarantee an >90% correctness rate for ORFs following sequencing of the 48 selected clones.
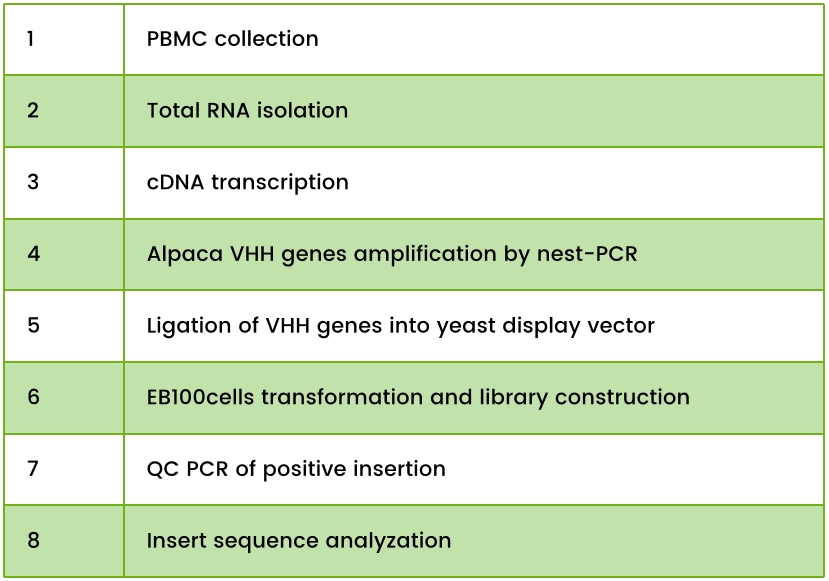
Table 1. Yeast Surface Display Library Construction Workflow Steps
Yeast Surface Display Screening Procedures
Iterative selection rounds utilize magnetic bead-based enrichment followed by flow cytometric sorting to isolate high-affinity, specific candidate clones. This dual-step process facilitates the identification of binder proteins with optimal binding properties and functional stability, providing a robust platform for generating lead candidates for downstream applications.

Fig 2. Workflow of yeast surface display screening
Features of Yeast Surface Display Platform
1. Provides a direct connection between genotype and phenotype.
2. Eukaryotic translation machinery offers expression in a similar fashion as mammalian cells.
3. No indication of amplification biases during antibody library screening.
4. Accurate control over selection parameters during FACS screening.
5. Real-time kinetic observations during the selection process.
6. Selected binders further analyzed by NGS significantly increase the number of identified target-specific antibodies.
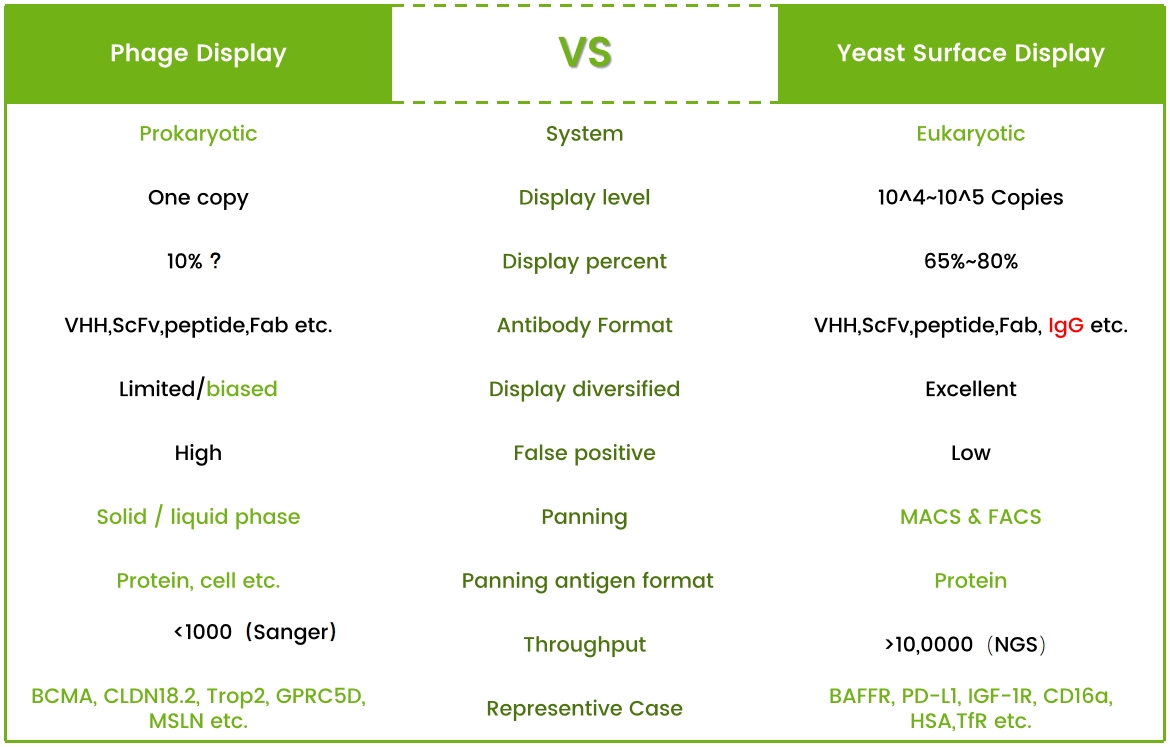
The difference between phage display and yeast display
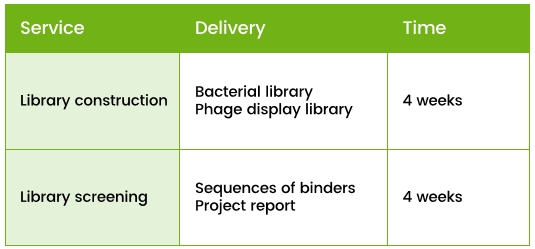
Service provided
Why chose our Yeast Surface Display system?
1. Stable platform with extensive experience.
2. High-quality yeast display library, with a capacity of up to 10^8 CFUs.
3. The library expression rates ranging from 70% to 80%, the highest industry standards.
4. Powerful combination of multi-color flow cytometry-based sorting and NGS sequencing.