
- Home
-
Services
 Leading molecular discovery services
Leading molecular discovery services
 Antibody Discover Service
Antibody Discover Service
 Technology platform
Technology platform
 Technology optimization service
Technology optimization service
- Products
- Resources
- Publication
- Contact Us
In vitro antibody selection platforms such as phage and yeast surface display has enabled the rapid selection of binders from diverse libraries.This, in turn, accelerates the discovery process of assist many research and development projects, including the creation of new drugs for clinical use.
Traditionally, antibody display libraries are analyzed by isolation of 1E2 –1E3 clones in combination with Sanger sequencing. The main limitation of the standard screening method is that, during the biopanning process, some binding clones become dominant, resulting in identification of alimited number of different candidates after screening. clones with superior affinity or with other prominent functional properties may be present at low frequency and may not be readily detectable.
An alternative approach is to combine yeast surface display with next-generation sequencing (NGS), which method massively increased the capacity to sequence millions of clones in a very fast and inexpensive way.
Yeast surface display is a powerful technique for screening large libraries of proteins or peptides. In this technique, the protein or peptide of interest is fused to the surface of a yeast cell. By coupling this technique with flow cytometry and NGS, it is possible to sequence millions of antibody clones in a single experiment. This allows for the rapid identification of high-affinity antibodies against a target antigen.
NBbiolab yeast surface display platform uses a general “a-agglutinin” display system,nanobodies display at the N-terminus of Aga2p, HA tag was fused to the C-terminus of the a-agglutinin Aga2p subunit. Following translation, the 69-amino acid Aga2p subunit associates with 725-amino acid a-agglutinin Aga1p subunit via two disulfide bonds. The fusion protein is subsequently secreted to the extracellular space where Aga1p is anchored to the cell wall via a β1,6-glucan covalent linkage. As a result, the naobodies displayed on the cell surface where it is accessible by soluble ligands.
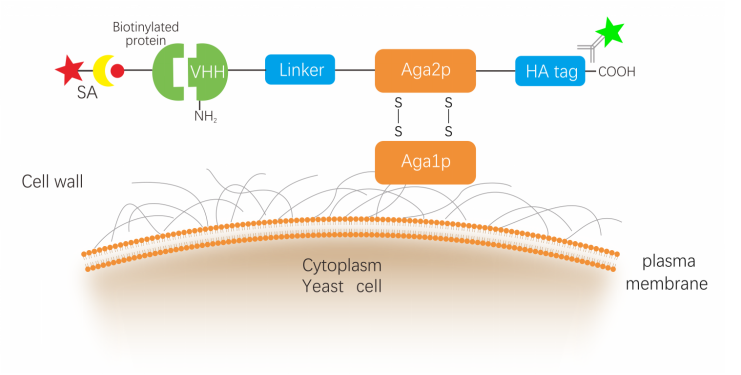
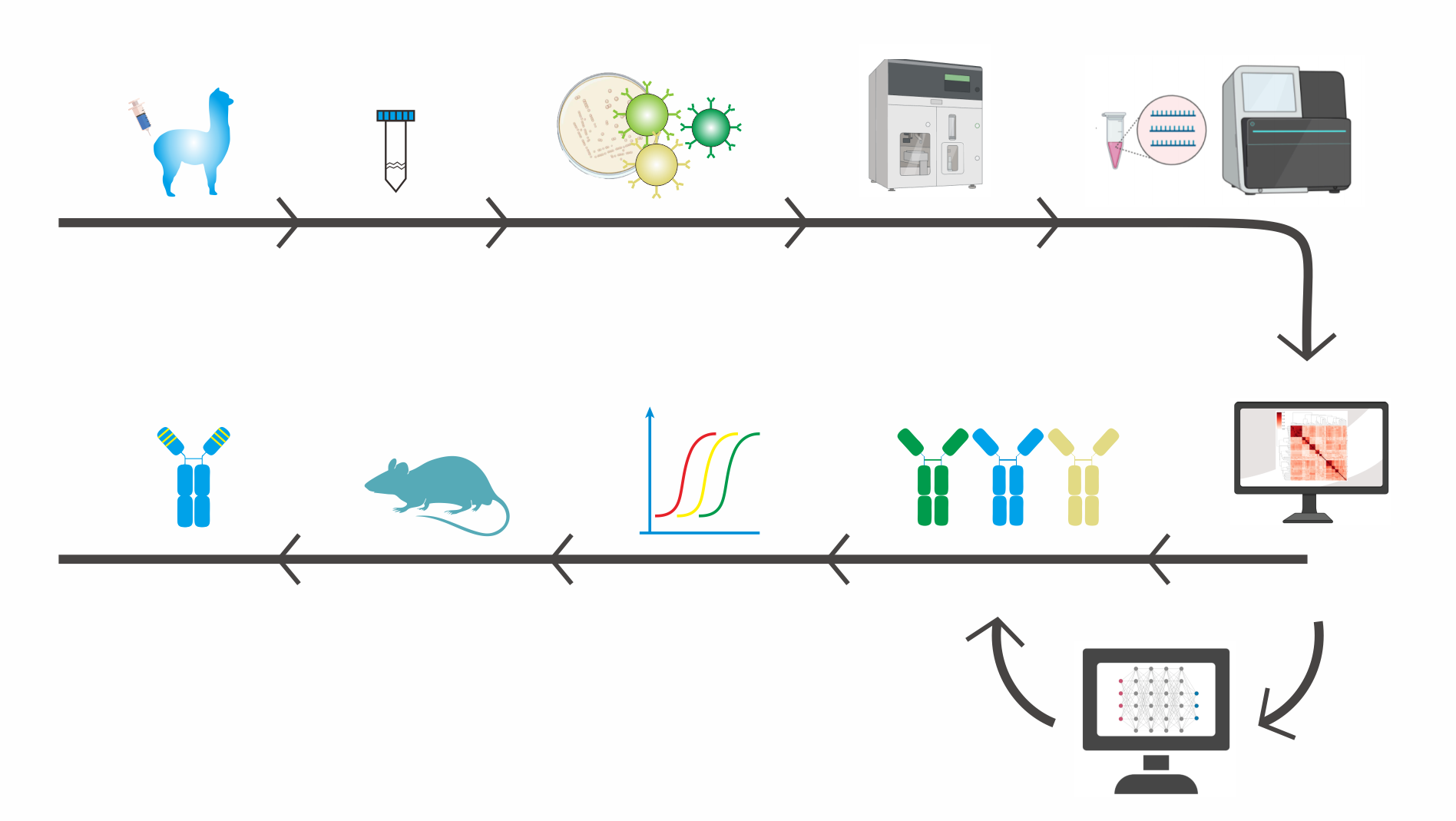
The general steps involved in yeast surface display screening
1. Create a yeast display library:
The first step is to generate a yeast library expressing the protein or peptide of interest fused to a surface protein, such as Aga2p or Aga1p. The library should be diverse enough to cover a wide range of potential binders.
2. Prepare the target molecule:
The target molecule can be a protein, peptide, small molecule or any other molecule of interest. The target molecule can be labeled with a fluorescent tag or biotin to allow for detection.
3. Bind the target molecule to the yeast library:
The yeast library is incubated with the target molecule to allow for binding. The unbound target molecules are then washed away.
4. Detect bound yeast cells:
The yeast cells that have bound to the target molecule can be detected using a fluorescent tag or biotin. If the target molecule is labeled with biotin, streptavidin can be used to detect the bound yeast cells.
5. Isolate bound yeast cells:
The yeast cells that have bound to the target molecule can be isolated using fluorescence-activated cell sorting (FACS) or magnetic-activated cell sorting (MACS).
6. Amplify and sequence bound yeast cells through NGS:
NGS approaches allow for far-greater insights into library diversity by providing up to 107 sequences, approximately 10,000-fold more sequences than Sanger sequencing.
7. Bioinformatic tools to analyze NGS Data:
Clone structures can be inferred by applying sequence clustering tools, to group closely related sequences into “clonal” groups. NGS data can also be used to guide selection toward novel sequence motifs.
8. Validate binding:
Once the candidate binders have been identified, the binding can be validated using additional assays, such as ELISA or surface plasmon resonance (SPR).
Features of Yeast Surface Display platform
1. Provides a direct connection between genotype and phenotype.
2. Eukaryotic translation machinery offers expression in a similar fashion as mammalian cells.
3. No indication of amplification biases during antibody library construction
4. Multivalent display with each cell displaying 104 to 105 copies of the VHH.
5. Accurate control over selection parameters during FACS screening.
6. Real-time kinetic observations during the selection process.
7. Selected binders further analyzed by NGS significantly increase the number of identified target-specific antibodies.