
- Home
-
Services
 Leading molecular discovery services
Leading molecular discovery services
 Antibody Discover Service
Antibody Discover Service
 Technology platform
Technology platform
 Technology optimization service
Technology optimization service
- Products
- Resources
- Publication
- Contact Us
CAR Single-domain Antibody Drug Development
CAR-T (Chimeric Antigen Receptor T-cell Therapy) is an innovative immunotherapy that targets tumor cells through CARs, triggering the release of cytokies to achieve potent anti-tumor effects. A CAR structure is primarily composed of three key domains:
· Extracellular Antigen-Binding Domain – Composed of antibody fragments (scFv or VHH) that determine antigen specificity and binding affinity, enabling precise tumor recognition.
· Transmembrane Domain – Anchors the receptor to the T-cell membrane, ensuring stability and facilitating signal transduction.
· Intracellular Signaling Domain – Contains co-stimulatory and signaling regions that drive T-cell activation, persistence, and tumor-killing potency.
The extracellular antibody fragment is crucial for CAR-T efficacy. High specificity minimizes off-target effects, while high affinity enhances antigen binding, improving tumor recognition and therapeutic outcomes. Optimizing CAR sequences is essential to balance these factors, maximizing tumor-killing efficiency while reducing cytokine release syndrome and other side effects.
Service Features
To support the development of high-quality CAR sequences, we offer two robust antibody discovery platforms:
1. Phage Display Platform
· Enables the identification of a diverse range of binding sequences.
2. Yeast Surface Display Platform
· Facilitates the discovery of antibody fragments with excellent binding characteristics.
Utilizing these advanced platforms, we can generate diverse binding sequences tailored to your therapeutic needs. By analyzing candidate molecules, we design multi-epitope or multi-specific CAR constructs, further enhancing the versatility and efficacy of your CAR-T therapies.
Service Process

Service Advantages

Case Studies
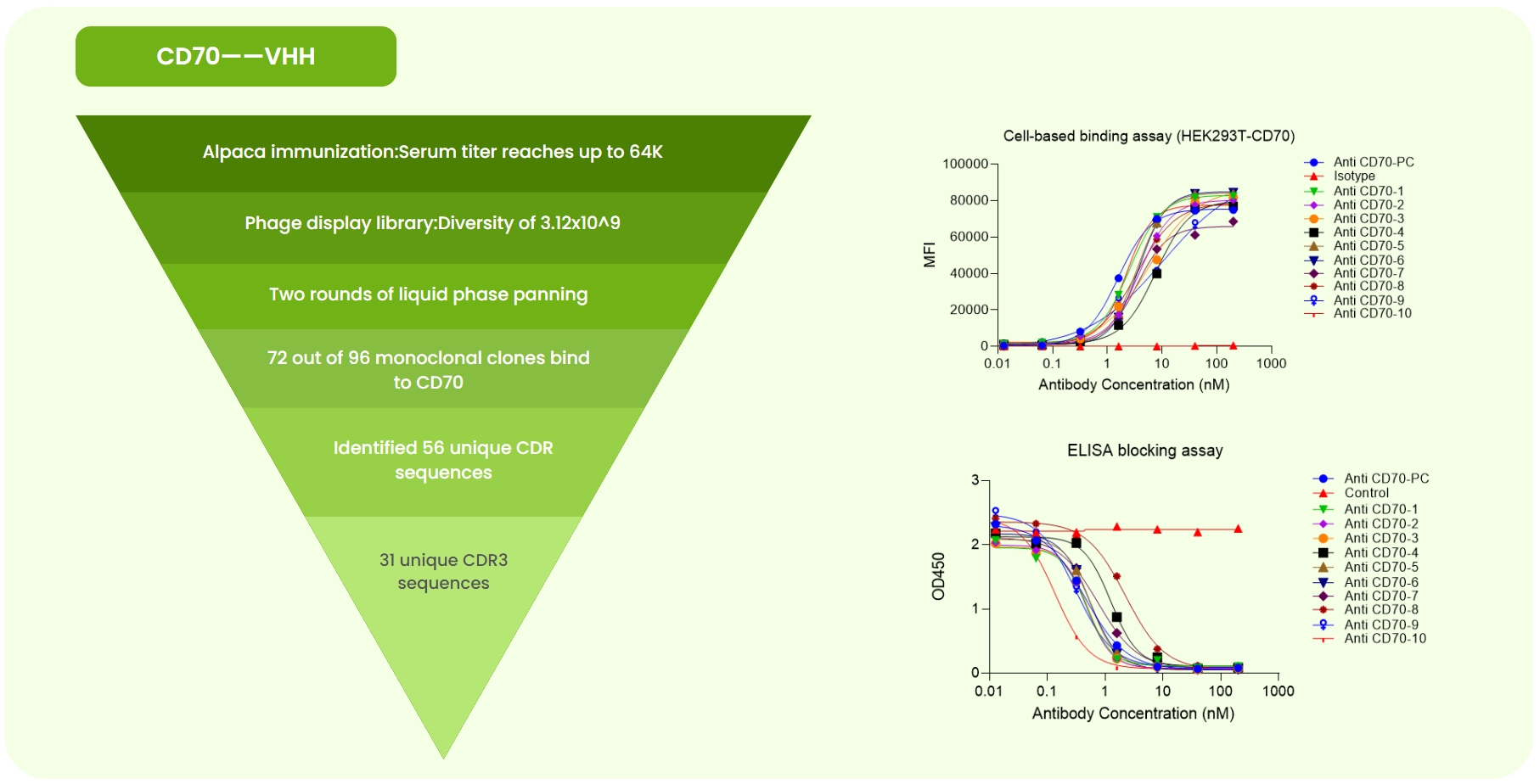
Fig 1. CD70 Single-Domain Antibody Development. (a) Immunized alpaca ELISA at 1:64K dilution showed OD >0.2. A 3.12×10⁹ phage library was constructed, followed by two rounds of liquid-phase panning. Sanger sequencing of 96 monoclones identified 56 unique CDR sequences. Expression and validation yielded 31 high-specificity antibodies. (b) Flow cytometry revealed varying binding strengths of anti-CD70-PC (benchmark), isotype, and 10 candidates to CD70-overexpressing cells. (c) ELISA confirmed different binding strengths of the same antibodies to CD70 protein.
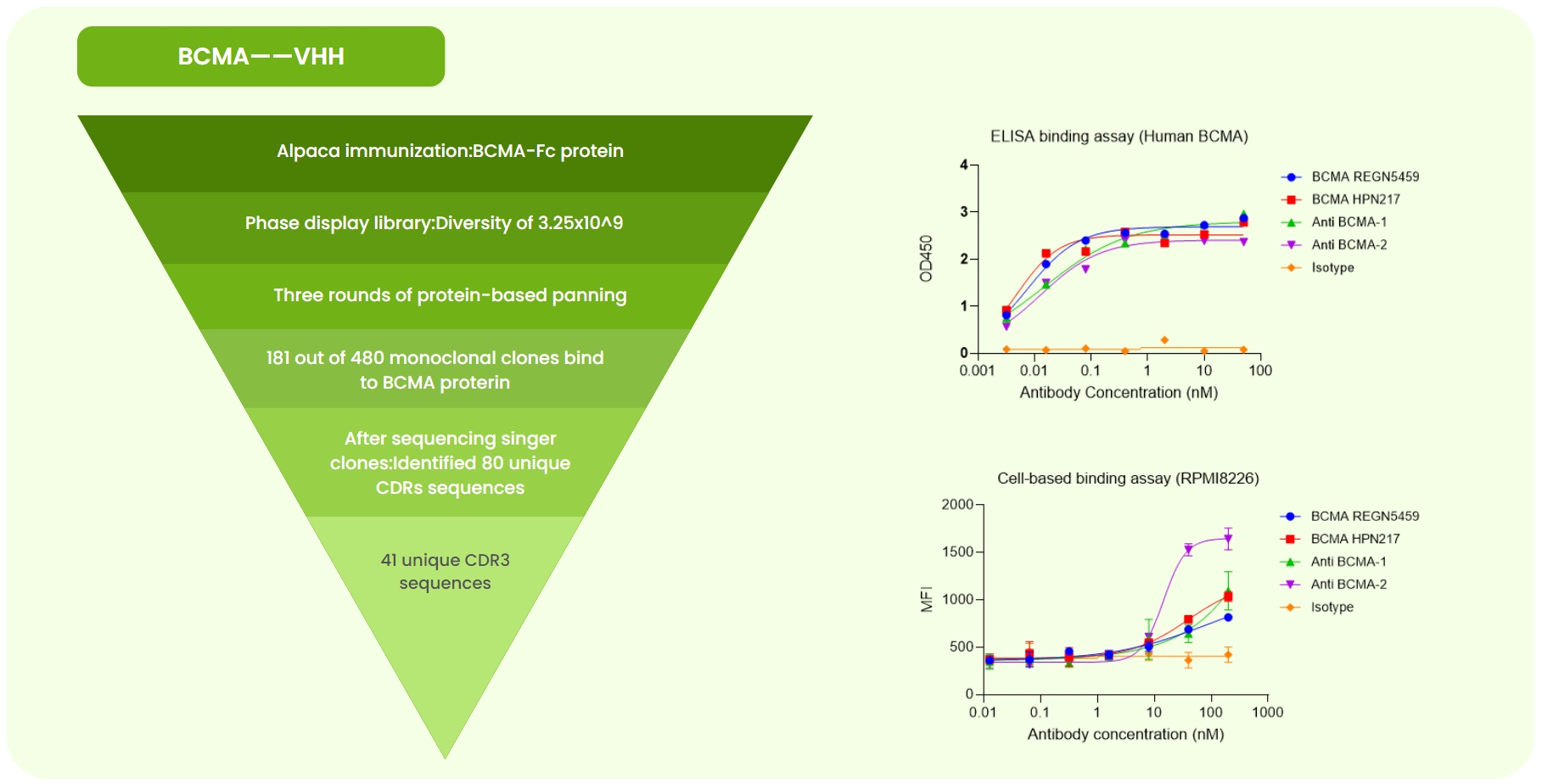
Fig 2. BCMA Single-Domain Antibody Development. (a) Alpaca immunized with BCMA-Fc protein. A 3.25×10⁹ phage library was constructed, followed by three rounds of liquid-phase panning. Sanger sequencing of 480 monoclones identified 80 unique CDR sequences. Expression and validation yielded 41 high-specificity antibodies. (b) ELISA detected varying binding strengths of BCMA REGN5459 (benchmark), BCMA HPN217 (benchmark), isotype, and two candidates to BCMA protein. (c) Flow cytometry showed that anti-BCMA-2 exhibited stronger binding to RPMI8226 cells than the benchmarks.
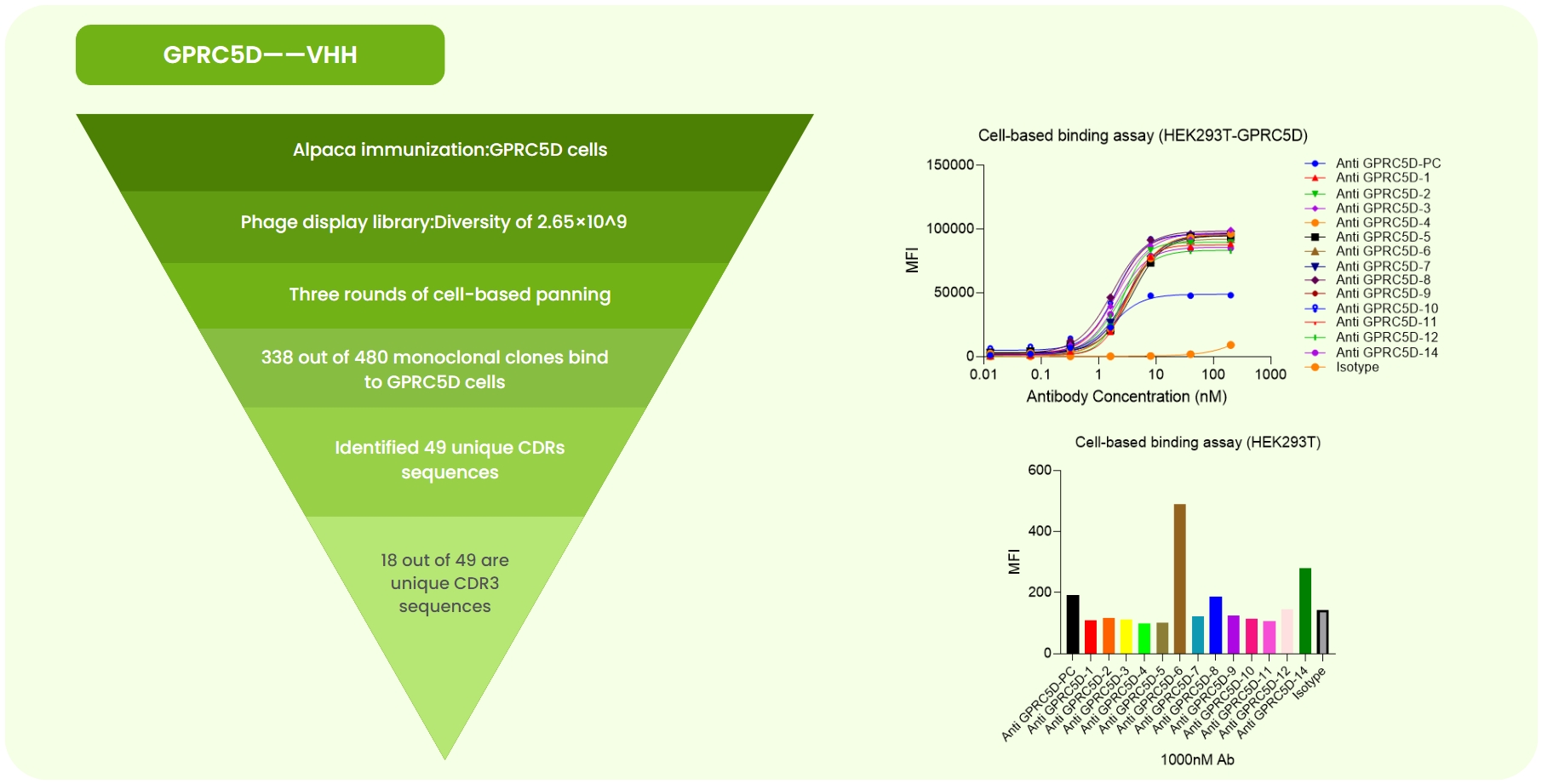
Fig 3. GPCR5D Single-Domain Antibody Development. (a) Alpaca immunized with GPCR5D-expressing cells. A 2.65×10⁹ phage library was constructed, followed by three rounds of cell panning. Sanger sequencing of 480 monoclones identified 49 unique CDR sequences. Expression and validation yielded 18 high-specificity antibodies. (b) Flow cytometry showed superior affinity of 13 candidates compared to the anti-GPCR5D-PC (benchmark) on GPCR5D-overexpressing cells. (c) At 100 nM, flow cytometry revealed varying binding strengths among candidates.